05 October 2020
Molecular signatures of beef tenderness: a robust list of biomarkers released
Ground breaking analysis has led to the publication of the world’s* most robust set of beef tenderness biomarkers and major molecular signatures of beef tenderness arising from research led by Dr. Mohammed Gagaoua, a Marie Skłodowska-Curie Career-FIT Fellow at the Food Quality and Sensory Science department at Teagasc, Ashtown.
Molecular signatures of beef tenderness: a robust list of biomarkers released
Many large omics datasets, especially proteomics, have been generated in the quest to identify molecular signatures for meat tenderness which in turn will lead to the development of a management system for guaranteed beef tenderness. Due to variations in experimental approaches e.g. cattle type, age at slaughter etc in generating these datasets, there is a need for a holistic and integrative study (integromics). An integromics approach is one which integrates and interrogates large and complex datasets. Employing an integromics approach, Dr Gagaoua’s publication is the first of its kind and has provided clarity on the most consistently and robustly identified biomarkers of beef tenderness. It gathers data from 28 independent proteomics-based experiments from which a comprehensive list of 124 biomarkers were initially identified. From these, the authors shortlisted a panel of 33 robust candidates worthy of evaluation using targeted or untargeted data-independent acquisition proteomic methods to develop future predictive tools. Furthermore, this research, as well as revealing the main molecular signatures, has also provided key insights into the interconnectedness among various pathways and processes in the muscle which are pivotal in producing high quality beef.
Dr Gagaoua highlights that this study has “revealed the relevance in the order of importance of muscle contractile and structure proteins, energy metabolism proteins, response to stress proteins and oxidative stress proteins in the determination of beef tenderness”. Interestingly, the authors revealed that most of these pathways and proteins directly or indirectly impinge on apoptosis onset in post-mortem muscle.
Dr Anne Maria Mullen, co-author and Teagasc Academic Mentor to Dr Gagaoua, stated that “while many advances have been realised in improving beef eating quality, unexplained variation remains. This integromics approach has been pivotal in identifying the most important and relevant molecular targets to advance our knowledge and lead to the development of systems for guaranteeing beef tenderness.”
Declan Troy, co-author and Assistant Director of Research in Teagasc, welcomes this research and highlights its value in bringing together vast omics studies in an applications-focused manner to address our gaps in knowledge and technology to greatly increase the efficiency of the beef industry in Ireland and on a global level. This study has the potential to be of tremendous value to the beef industry worldwide and more specifically for Irish beef industry.
“This work would not be possible without the support of Marie Skłodowska-Curie grant agreement No. 713654 and that of Meat Technology Ireland a co-funded industry/Enterprise Ireland project”, says the first author Mohammed Gagaoua.
* The publication on the biomarkers of beef tenderness was published in Meat Science by a team of researchers at Teagasc and Meat Technology Ireland in collaboration with prominent meat scientists from INRAE (France), Centro Tecnológico de la Carne de Galicia (Spain), University of Melbourne (Australia), Centro de Investigacion Veterinaria de Tandil (Argentina), Virginia Tech (USA) and NSW DPI, Centre for Red Meat and Sheep Development (Australia).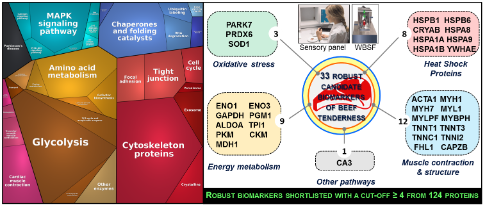
Full reference: Gagaoua M., Terlouw E.M.C., Mullen A.M., Franco D., Warner R.D., Lorenzo J.M., Purslow P.P., Gerrard D., Hopkins D.L., Troy D. & Picard B. (2021). Molecular signatures of beef tenderness: Underlying mechanisms based on integromics of protein biomarkers from multi-platform proteomics studies. Meat Sci 172, 108311. https://doi.org/10.1016/j.meatsci.2020.108311
